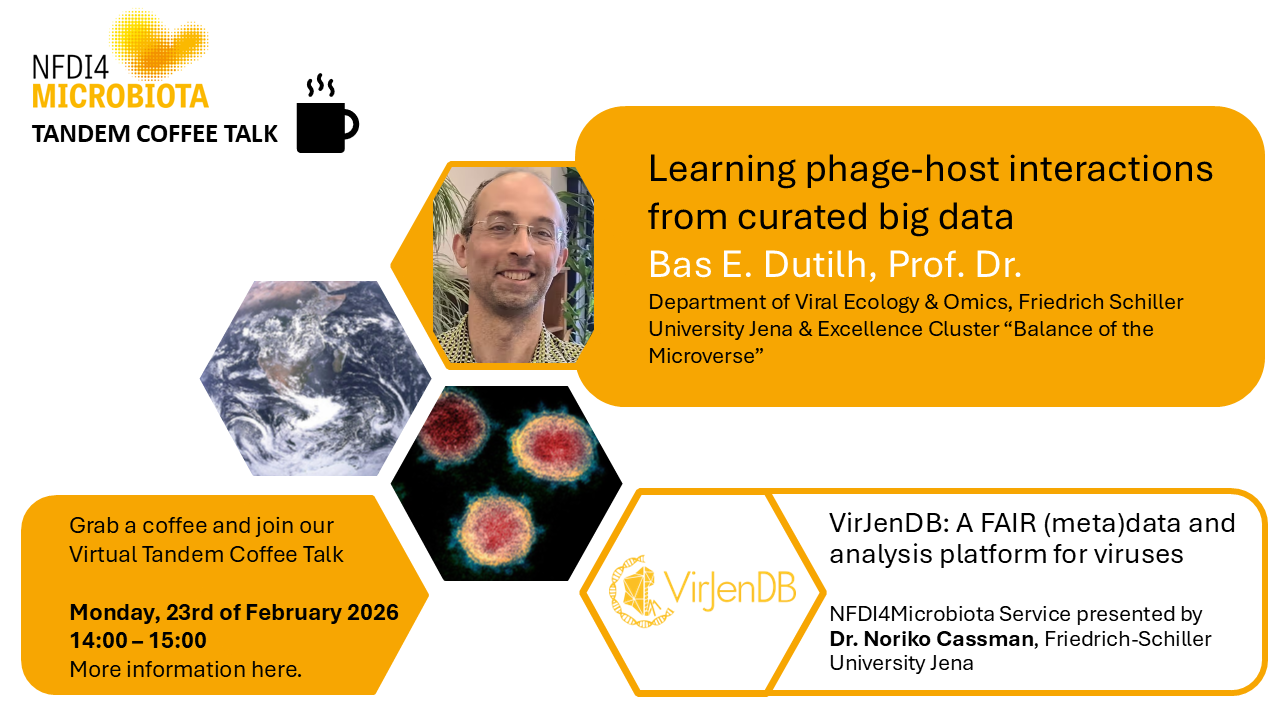
Making Microbiology Data FAIR and Open
As a part of the Nationale Forschungsdaten Infrastruktur (NFDI), NFDI4Microbiota supports the microbiology community with access to data, analysis services, data/metadata standards, and training.
What we offer
Our consortium offers a wide range of services, knowledge and best practice. Below you find the main topics on which we support our community.
jason-goodman-unsplash.jpg)
john-schnobrich-unsplash.jpg)
christopher-gower-unsplash.jpg)
markus-spiske-unsplash.jpg)
marvin-meyer-unsplash.jpg)
arlington-research-unsplash.jpg)