TAXA
Descriptions of novel taxonomic lineages
Short description
A substantial fraction of bacteria are unknown and undescribed, but as genomes exist for these unknown taxa, it is possible to describe them using genome-inferred information. However, the description of novel taxa currently lacks standardized formats and there is limited quality assurance during this process.
TAXA will fill this gap. The idea behind it is to use the published bioinformatic tool Protologger to provide standardized functional and taxonomic readouts of novel taxa using genome sequences, which will provide the building blocks for the community to describe them. This information will be provided to the community via a custom Wiki, allowing members of the community to contribute by converting the building blocks we provide into fully formed taxonomic descriptions. This tackles the problem of ambiguous naming practices in the process of validly describing novel species. Via the Wiki, researchers will be able to identify interesting and relevant bacteria to their research question at a glance.
Graphical abstract
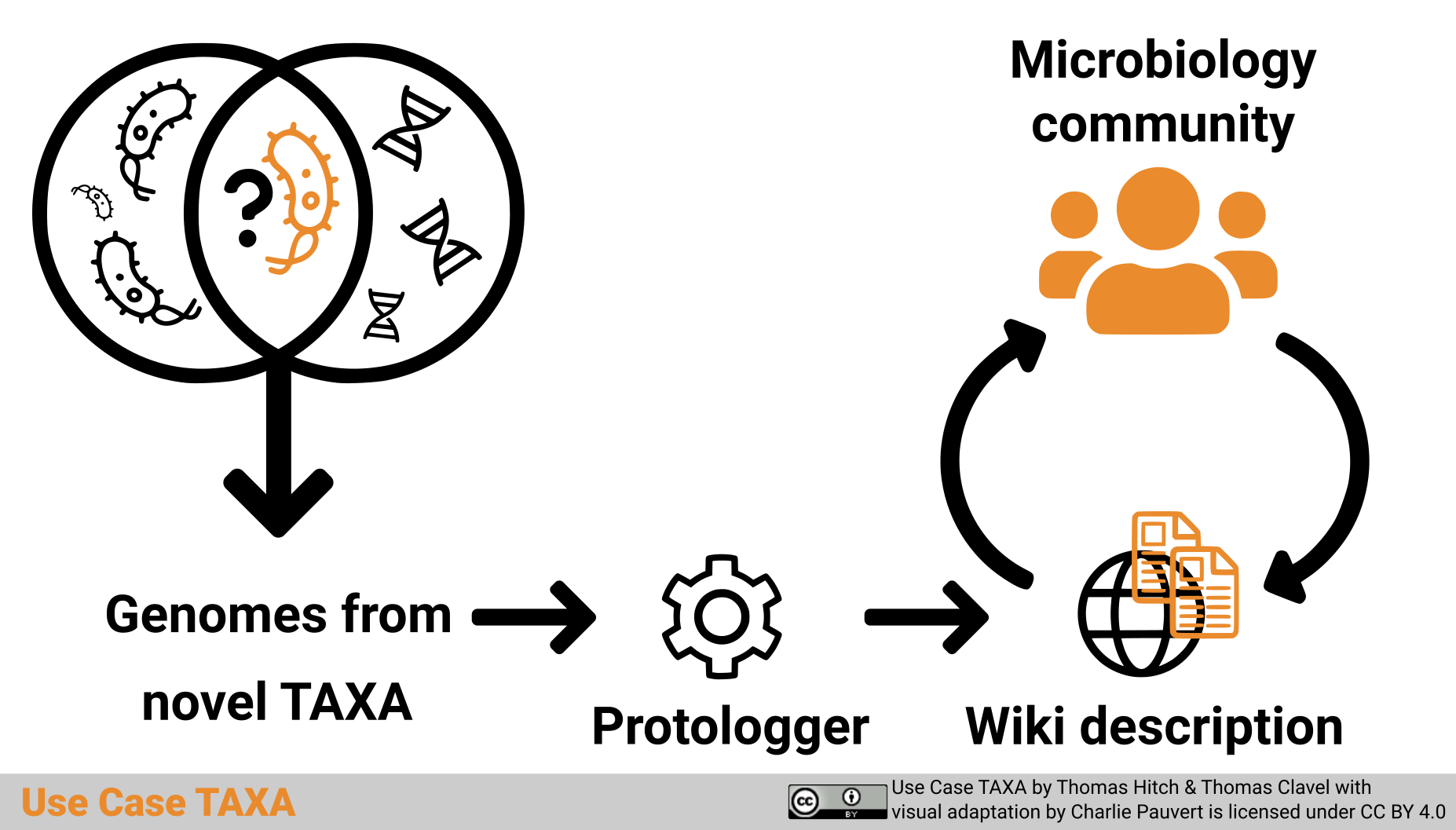
Graphical abstract “Use Case TAXA” by Thomas Hitch and Thomas Clavel with visual adaptation by Charlie Pauvert is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0).
How you can contribute
You are a …
- microbiologist with undescribed novel species:
Get trained on using the tool and generate standardized descriptions of bacteria by using it - metagenome analyst frustrated with numerical bacteria names lacking functional insights:
Use the tool to generate functional descriptions and curate information in a human-readable format - computation scientist:
Enhance the capacities of our tool by interacting with the team of developers
Planned output
Standardization through automated pipeline
- Improve the existing easy tool for taxa description under standardized conditions using sequence-based analysis of microbiome species
Standardized output
- Using the integrated analysis pipeline to provide data output via a user-friendly interface in form of a wiki template
- Generating standardized protologues for over 10,000 uncultured species (i.e. from deposited genomes in GTDB)
-> for human gut microbiomes as a start
-> then expand to other ecosystems like soil and marine environments - Create FAIR datasets by including linked open metadata on a WikiBase instance in collaboration with the Use Case MicroBioKGs
Human-readable output
- Converting data output into human-readable descriptions of novel taxa
How to jointly achieve all output
- Using the analysis pipeline already available for all interested researchers
- Making the pipeline applicable for other disciplines by enabling scalability (Docker instance)
- Establishing training videos for researchers on using the pipeline
- Standardizing taxonomic descriptions via the interactive interface of the wiki template
- Promoting high-quality taxonomic descriptions of bacteria by the community via the wiki template
Achievements
- Updated Protologger and transferred service to de.NBI servers
Project Lead

Thomas Hitch
ORCID ID: 0000-0003-2244-7412
Institute of Medical Microbiolgy, Junior Group, Research Group Functional Microbiome,University Hospital of RWTH Aachen

Thomas Clavel
ORCID ID: 0000-0002-7229-5595
Institute of Medical Microbiolgy, Head of Research Group Functional Microbiome, University Hospital of RWTH Aachen
Keywords
microbiomes
taxonomy
novel bacterial species
nomenclature
protologues
bioinformatic tool
crowd-based