VirJenDB
Curated virus database
Short description
The VirJenDB (VJDB) is a virus database currently being developed in Jena, Germany.
As a Use Case and service of the NFDI4Microbiota, the VJDB combines the elements of the available NFDI infrastructure - the de.NBI Cloud, CLoWM workflow system and Aruna Object Storage - to create a cloud-based solution for the re-use of virus data in line with the FAIR and Open Science principles.
The VJDB features a web interface for researchers to search, browse, and download an initial dataset of sequences and metadata from the ENA, NCBI Virus and other repositories.
Plans to expand the platform include adding automated and community-curated analysis workflows, additional data types including imaging data, and visualization and sharing options.
The VJDB aims to be a community-led platform featuring user-friendly tools for all viruses and incorporating the latest developments from the infrastructure and database fields.
Graphical abstract
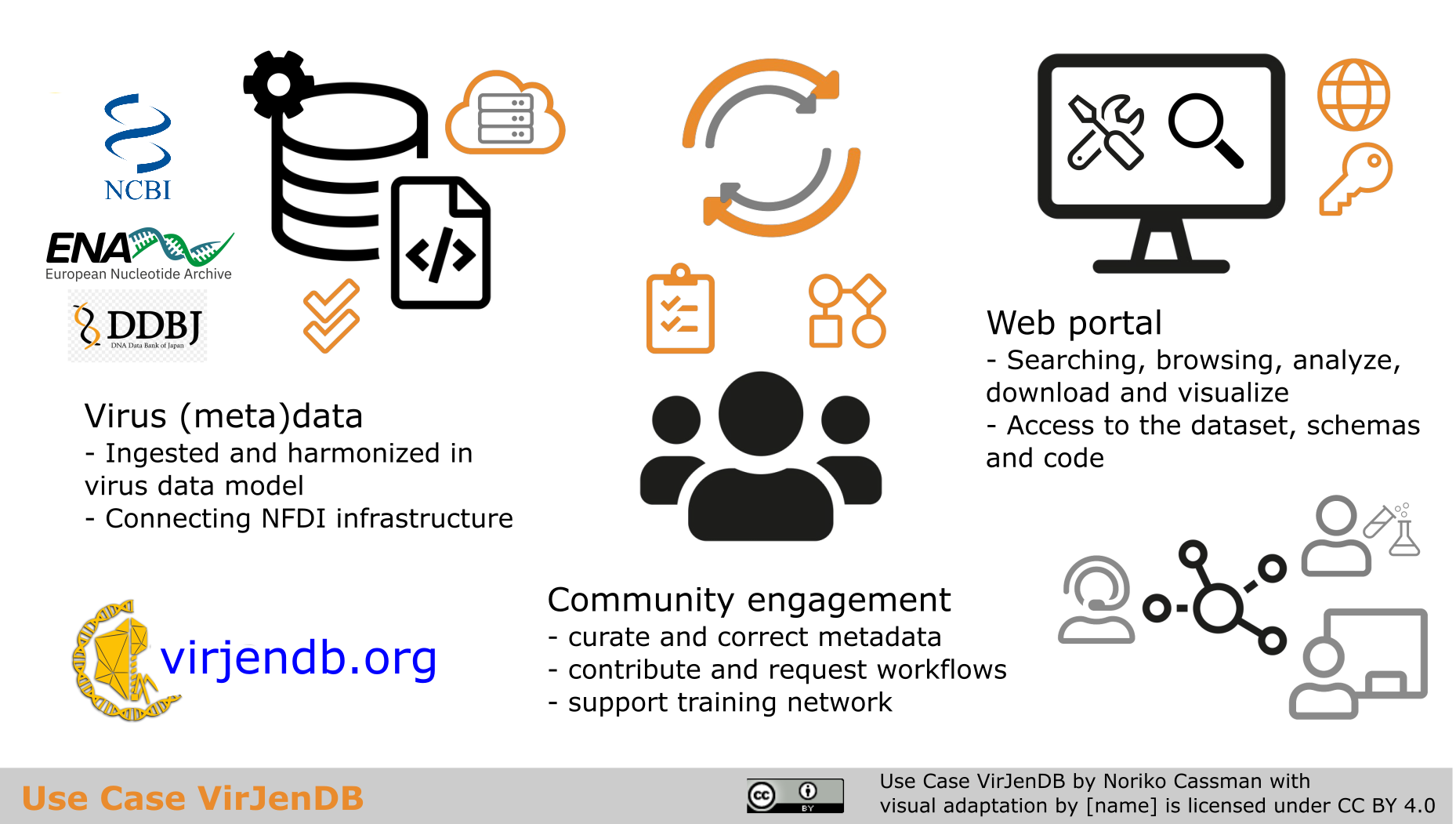
Graphical abstract “Use Case VirJenDB” by Noriko Cassman is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0).
How you can contribute
You are a …
- (non-bioinformatician) virologist with not yet published virus genome sequences:
Get support to submit your sequences to the ENA for publication - (non-bioinformatician) virologist with published data in ENA:
Get further trained in RDM topics and contribute to community-curated datasets and resources - (non-bioinformatician) virologist with a need for preliminary data analysis results, e.g., to include in a proposal:
Use the suggested tools (and get trained) to quickly generate preliminary results to see a first trend to answer your research question - (bioinformatician) phage ecologist looking for data to include in your analysis pipeline:
Use the VJDB API and interface to get data (e.g., ready-to-analyze sets of sequence data) - (bioinformatician) phage ecologist with limited financial resources:
Use our easy-to-use and ready-made analysis and infrastructure capabilities to your advantage
Planned output
Research output
- Developing a virus metadata model and contributing to existing metadata standards
- Releasing a versioned VJDB virus data model regularly (currently the v0.1 is available)
- Release validation parameters and standard suggestions
- Developing the VJDB dataset with regular updates (currently the beta version 0.1 is available)
- Listed virus resources via the NFDI4Microbiota Knowledge Base
- Accessing through the web interface (search, browse, visualize and download predefined subsets)
- Provide bulk downloads via an API
Infrastructure output
- Integrate modular, open-source bioinformatics workflows and custom pipelines from the de.NBI, CloWM system and others
- Accessing all code and metadata schemas via Github and adding relations to CLoWM system
- Connecting users to existing ENA submission tool to prompt sequence and metadata submission to ENA
- Update researcher’s own erroneous metadata and sequences in the ENA via a curation workflow
- Integrate sequence search, assembly, clustering, gene prediction and annotation workflows
- Integrate a (meta)genome and structural viewer and a private user workbench
- Training researchers on the use of the VJDB and increasing knowledge on available virus research resources
- Seeking out researchers to contribute expertise by networking and demos at relevant conferences and events
- Helping to organize relevant and related workshops
- Create tutorials for beginner, intermediate and advanced users
Achievements
Project Lead
Noriko Cassman
ORCID ID: 0000-0003-1655-0931
Friedrich Schiller University Jena

Manja Marz
ORCID ID: 0000-0003-4783-8823
Friedrich Schiller University Jena
Shahram Saghaei
ORCID ID: 0009-0005-5554-275X
Friedrich Schiller University Jena
Keywords
virus
sequence
metadata
database
research infrastructure