MetaProt
Functional profiling and routine diagnosis of humane microbiomes by metaproteomics
Short description
Metaproteomics is a powerful method to acquire in-depth data on microbiomes, with the human gut microbiome being of particular interest to researchers. LC-MS/MS-based metaproteomics is used to characterize taxonomic and functional microbiome composition in the field of health, environmental sciences, and biotechnology. The abundance of proteins generally correlates well with actual activity and can detect proteins from the environment, such as host protein in the human gut or substrate proteins in biogas plants. The long-term goal of the metaproteomics research community is to contribute to the complete exploration of the human gut microbiome and to offer an attractive option for routine diagnostics. Standardization and data integration are key milestones toward this goal. Routine diagnostics is only feasible if a large enough body of data is available. In principle, this data already exists, but further efforts of data integration have to be undertaken.
The Use Case MetaProt aims at standardization and data integration in human gut metaproteomics by achieving two objectives:
- the creation of a curated protein sequence database of the human gut microbiome, and
- the creation of a metadata annotation and file conversion tool that produces a uniform output from diverse sources of experimental data.
Graphical abstract
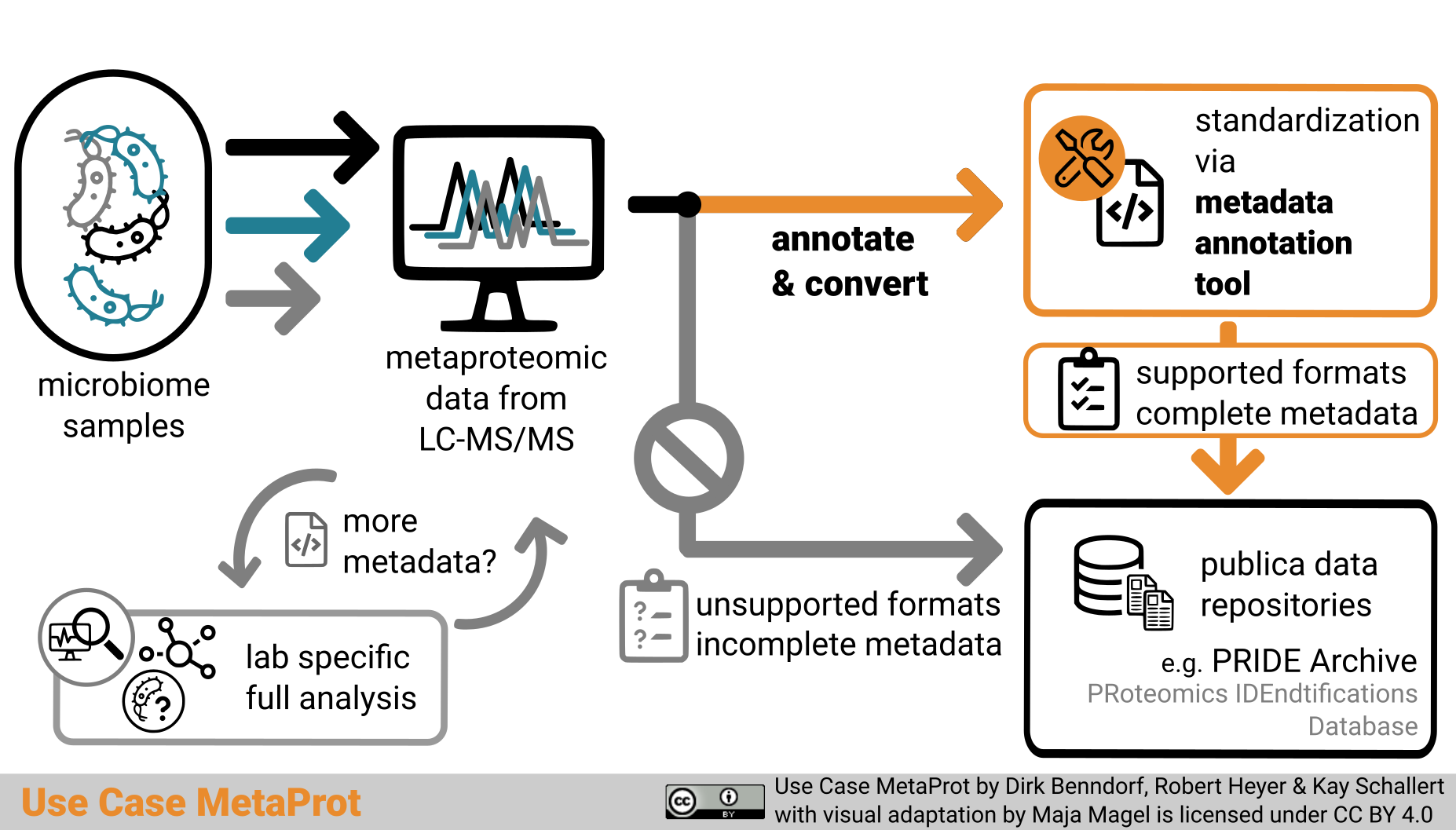
Graphical abstract “Use Case MetaProt” by Dirk Benndorf, Robert Heyer and Kay Schallert with visual adaptation by Maja Magel is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0).
How you can contribute
You are a …
- microbiologist beginner interested in metaproteomics:
Get trained on metaproteomics data handling focusing on the bioinformatics aspect, and the tool MetaProteomeAnalyzer. - microbiologist with knowledge of omics metadata standards:
Reach out to us during conferences and contribute to standardizing metadata collection across multiple omics communities - bioinformatician curating metagenomes datasets:
Provide curated data and metadata from human gut microbiome samples.
Planned output
- Establish requirements for metaproteomics data handling and establish a metadata standard
- Discuss tool and standard compatibility with other omics communities
- Develop standardized database for protein identification based on existing protein sequence databases
- Present and advocate for the use of the metadata annotation tool on conferences
- Hosting a workshop to teach metadata annotation to researchers in the field
Research output
- Publication about metaproteomics metadata standard and best practices
Infrastructure output
- Web service for organizing metaproteomics metadata
- Web page providing databases for protein identifications in metaproteomics
Achievements
- Delivered hands-on workshop on Applied Metaproteomics in 2023
- User friendly interface for demo web services MetaForge and MPA: https://mdoa-tools.bi.denbi.de/home
Project Lead
Dirk Benndorf
ORCID ID: --
Applied Biosciences and Process Engineering, Anhalt University of Applied Sciences, Microbiology, Bernburger Straße 55, 06354 Köthen

Robert Heyer
ORCID ID: --
(1) Multidimensional Omics Data Analysis group, Leibniz-Institut für Analytische Wissenschaften – ISAS – e.V., Bunsen-Kirchhoff-Straße 11, 44139 Dortmund;
(2) Faculty of Technology, Bielefeld University, Universitätsstraße 25, 33615 Bielefeld
Keywords
metaproteomics
gut microbiome
routine diagnostics
health care
systems biology